Note
Go to the end to download the full example code. or to run this example in your browser via Binder
Landmark shift#
This example shows how to shift functional data objects to align its samples with a particular reference point.
# Author: Pablo Marcos Manchón
# License: MIT
# sphinx_gallery_thumbnail_number = 2
We will use an example dataset synthetically generated by
make_multimodal_samples(), which in this case will be
used to generate gaussian-like samples with a mode near to 0.
Each sample will be shifted to align their modes to a reference point using
the function
landmark_shift_registration().
import matplotlib.pyplot as plt
from skfda.datasets import make_multimodal_samples
fd = make_multimodal_samples(random_state=1)
fd.extrapolation = "bounds" # See extrapolation for a detailed explanation.
fd.plot()
plt.show()

A landmark or a feature of a curve is some characteristic that one can associate with a specific argument value t. These are typically maxima, minima, or zero crossings of curves, and may be identified at the level of some derivatives as well as at the level of the curves themselves[1].
For alignment we need to know in advance the location of the landmark of
each of the samples, in our case it will correspond to the maxima of each
sample. Because our dataset has been generated synthetically we can obtain
the value of the landmarks using the function
make_multimodal_landmarks(), which is used by
make_multimodal_samples() to set the location of the
modes.
In general it will be necessary to use numerical or other methods to determine the location of the landmarks.
import numpy as np
from skfda.datasets import make_multimodal_landmarks
landmarks = make_multimodal_landmarks(random_state=1).squeeze()
fig, ax = plt.subplots()
ax.scatter(landmarks, np.repeat(1, fd.n_samples))
fd.plot(fig=fig)
plt.show()
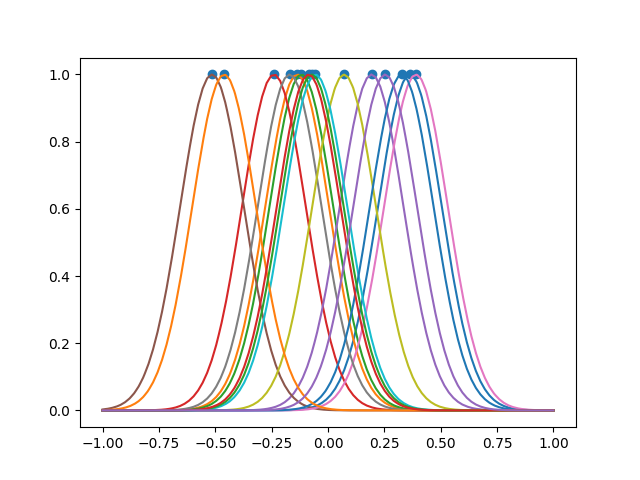
Location of the landmarks:
print(landmarks)
[ 0.36321467 -0.13679289 -0.11810279 -0.23992308 0.19351103 -0.5146397
0.39015177 -0.17021104 0.07133931 -0.05576091 0.32693727 -0.46066147
-0.07209468 -0.08587716 0.25351855]
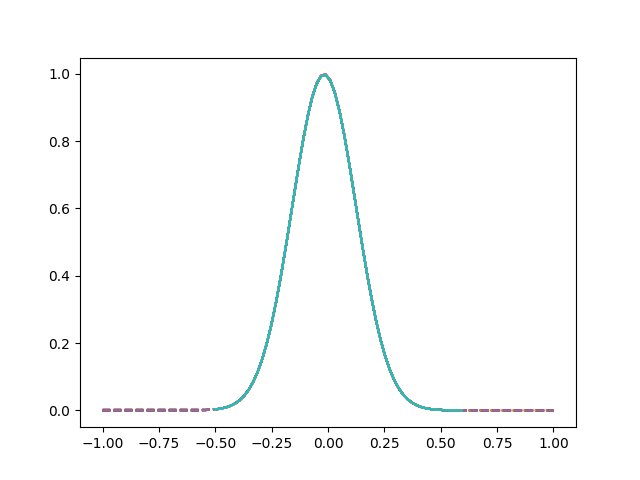
The following figure shows the result of shifting the curves to align their landmarks at 0.
from skfda.preprocessing.registration import landmark_shift_registration
fd_registered = landmark_shift_registration(
fd,
landmarks,
location=0,
)
fig = fd_registered.plot()
fig.axes[0].scatter(0, 1)
plt.show()

In many circumstances it is possible that we could not apply extrapolation, in these cases it is possible to restrict the domain to avoid evaluating points outside where our curves are defined.
If the location of the new reference point is not specified it is choosen the point that minimizes the maximum amount of shift.
# Curves aligned restricting the domain
fd_restricted = landmark_shift_registration(
fd,
landmarks,
restrict_domain=True,
)
# Curves aligned to default point without restrict domain
fd_extrapolated = landmark_shift_registration(
fd,
landmarks,
)
fig = fd_extrapolated.plot(linestyle="dashed", label="Extrapolated samples")
fd_restricted.plot(fig=fig, label="Restricted samples")
plt.show()
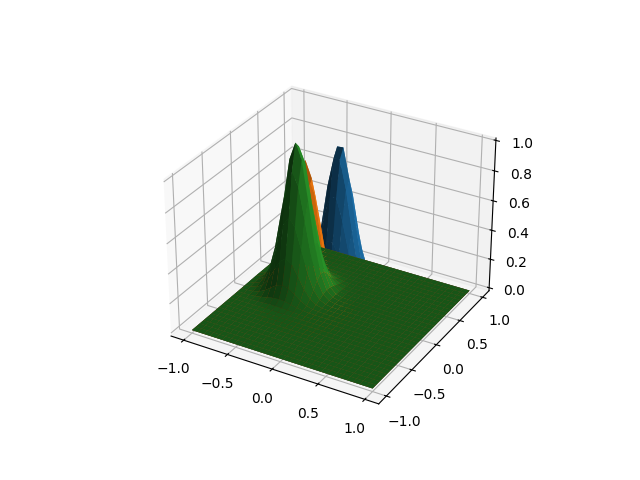
The previous method is also applicable for multidimensional objects, without limitation of the domain or image dimension. As an example we are going to create a datset with surfaces, in a similar way to the previous case.
fd = make_multimodal_samples(
n_samples=3,
points_per_dim=30,
dim_domain=2,
random_state=1,
)
fd.plot()
plt.show()
In this case the landmarks will be defined by tuples with 2 coordinates.
landmarks = make_multimodal_landmarks(
n_samples=3,
dim_domain=2,
random_state=1,
).squeeze()
print(landmarks)
[[ 0.36321467 -0.13679289]
[-0.11810279 -0.23992308]
[ 0.19351103 -0.5146397 ]]
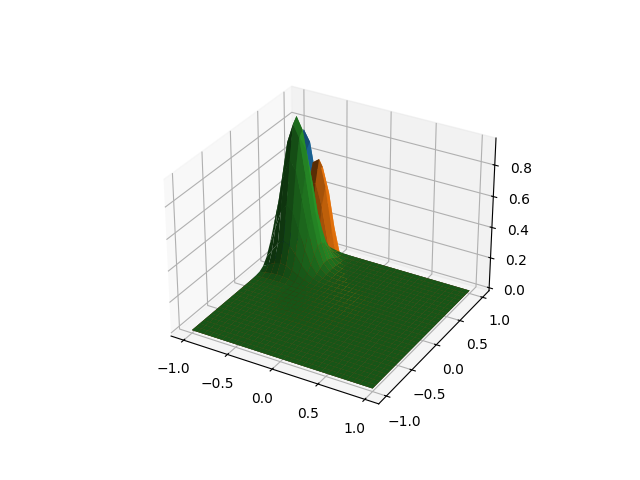
As in the previous case, we can align the curves to a specific point, or by default will be chosen the point that minimizes the maximum amount of displacement.
fd_registered = landmark_shift_registration(
fd,
landmarks,
)
fd_registered.plot()
plt.show()
Total running time of the script: (0 minutes 0.492 seconds)
