Note
Go to the end to download the full example code. or to run this example in your browser via Binder
Clustering#
In this example, the use of the clustering plot methods is shown applied to the Canadian Weather dataset. K-Means and Fuzzy K-Means algorithms are employed to calculate the results plotted.
# Author: Amanda Hernando Bernabé
# License: MIT
# sphinx_gallery_thumbnail_number = 6
First, the Canadian Weather dataset is downloaded from the package ‘fda’ in CRAN. It contains a FDataGrid with daily temperatures and precipitations, that is, it has a 2-dimensional image. We are interested only in the daily average temperatures, so we select the first coordinate function.
import numpy as np
from skfda.datasets import fetch_weather
X, y = fetch_weather(return_X_y=True, as_frame=True)
fd = X.iloc[:, 0].array
target = y.array
fd_temperatures = fd.coordinates[0]
# The desired FDataGrid only contains 10 random samples, so that the example
# provides clearer plots.
indices_samples = np.array([1, 3, 5, 10, 14, 17, 21, 25, 27, 30])
fd = fd_temperatures[indices_samples]
The data is plotted to show the curves we are working with. They are divided according to the target. In this case, it includes the different climates to which the weather stations belong to.
import matplotlib.pyplot as plt
climates = target[indices_samples].remove_unused_categories()
fd.plot(
group=climates.codes,
group_names=climates.categories,
)
plt.show()

The number of clusters is set with the number of climates, in order to see the performance of the clustering methods, and the seed is set to one in order to obatain always the same result for the example.
n_clusters = len(climates.categories)
seed = 2
First, the class KMeans is instantiated with
the desired. parameters. Its fit() method
is called, resulting in the calculation of several attributes which include
among others, the the number of cluster each sample belongs to (labels), and
the centroids of each cluster. The labels are obtaiined calling the method
predict().
from skfda.ml.clustering import KMeans
kmeans = KMeans(n_clusters=n_clusters, random_state=seed)
kmeans.fit(fd)
print(kmeans.predict(fd))
[0 1 0 0 0 2 2 1 0 2]
To see the information in a graphic way, we can use the class
ClusterPlot.
from skfda.exploratory.visualization.clustering import ClusterPlot
# Customization of cluster colors and labels in order to match the first image
# of raw data.
cluster_colors = ["C0", "C2", "C1"]
cluster_labels = climates.categories[np.array([0, 2, 1])]
ClusterPlot(
kmeans,
fd,
cluster_colors=cluster_colors,
cluster_labels=cluster_labels,
).plot()
plt.show()

Other clustering algorithm implemented is the Fuzzy K-Means found in the
class FuzzyCMeans. Following the
above procedure, an object of this type is instantiated with the desired
data and then, the
fit() method is called.
Internally, the attribute membership_degree_ is calculated, which
contains n_clusters elements for each sample and dimension, denoting the
degree of membership of each sample to each cluster. They are obtained
calling the method predict_proba().
Also, the centroids of each cluster are obtained.
from skfda.ml.clustering import FuzzyCMeans
fuzzy_kmeans = FuzzyCMeans(n_clusters=n_clusters, random_state=seed)
fuzzy_kmeans.fit(fd)
print(fuzzy_kmeans.predict_proba(fd))
[[0.8721254 0.11189295 0.01598165]
[0.4615364 0.51285956 0.02560405]
[0.97428363 0.01882257 0.0068938 ]
[0.91184323 0.05369029 0.03446648]
[0.79072268 0.18411219 0.02516513]
[0.178624 0.05881132 0.76256468]
[0.01099498 0.00492593 0.98407909]
[0.03156897 0.96349997 0.00493106]
[0.8084018 0.13418057 0.05741763]
[0.03767122 0.01891178 0.943417 ]]
To see the information in a graphic way, the class
ClusterPlot can
be used. It assigns each sample to the cluster whose membership value is the
greatest.
ClusterPlot(
fuzzy_kmeans,
fd,
cluster_colors=cluster_colors,
cluster_labels=cluster_labels,
).plot()
plt.show()

Another plot implemented to show the results in the class
FuzzyCMeans is
ClusterMembershipLinesPlot.
which is similar to parallel coordinates. It is recommended to assign colors
to each of the samples in order to identify them. In this example, the
colors are the ones of the first plot, dividing the samples by climate.
from skfda.exploratory.visualization.clustering import (
ClusterMembershipLinesPlot,
)
colors_by_climate = np.array(["C0", "C1", "C2"])[climates.codes]
ClusterMembershipLinesPlot(
fuzzy_kmeans,
fd,
cluster_labels=cluster_labels,
sample_colors=colors_by_climate,
).plot()
plt.show()

Finally, the class
ClusterMembershipPlot
has a plot method which
returns a barplot. Each sample is designated with a bar which is filled
proportionally to the membership values with the color of each cluster.
from skfda.exploratory.visualization.clustering import ClusterMembershipPlot
ClusterMembershipPlot(
fuzzy_kmeans,
fd,
cluster_colors=cluster_colors,
cluster_labels=cluster_labels,
).plot()
plt.show()
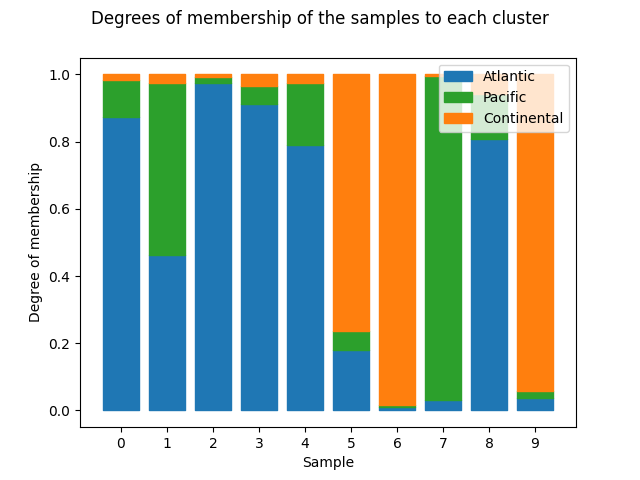
The possibility of sorting the bars according to a cluster is given specifying the number of cluster, which belongs to the interval [0, n_clusters).
We can order the data using the first cluster:
ClusterMembershipPlot(
fuzzy_kmeans,
fd,
sort=0,
cluster_colors=cluster_colors,
cluster_labels=cluster_labels,
).plot()
plt.show()

Using the second cluster:
ClusterMembershipPlot(
fuzzy_kmeans,
fd,
sort=1,
cluster_colors=cluster_colors,
cluster_labels=cluster_labels,
).plot()
plt.show()

And using the third cluster:
ClusterMembershipPlot(
fuzzy_kmeans,
fd,
sort=2,
cluster_colors=cluster_colors,
cluster_labels=cluster_labels,
).plot()
plt.show()

Total running time of the script: (0 minutes 0.569 seconds)
